Bobcat TAILS
Technology and Artificial Intelligence Learning Seminar
The Technology and Artificial Intelligence Learning Seminar (TAILS) is a weekly seminar on signals, dynamics, statistics, and machine learning as a part of the NSF AI Institute in Dynamic Systems and the Rocky Mountain Data Science student organization.
The seminar is open to any interested undergraduate and graduate students, as well as other researchers or faculty; Since this seminar is intended to be maximally inclusive, no particular prerequisites are expected other than healthy curiosity and a desire to learn concepts, tools, and tricks regarding data science. The goal of this long-term seminar is to train a cohort of "students" who will be well-equipped to tackle interesting applied and theoretical research questions in the realm of "signals and statistics" at large (audio, image, other data).
The seminar began with basic introductions of concepts but is transitioning to more interactive hands-on learning surrounding actual data, research, and results. Last summer, a number of graduate and undergraduate students undertook research relevant to TAILS topics. While we have specific topics, data, and research questions in mind, additional input is always welcome.
TAILS is particularly targeted towards undergraduates who are interested in "data science research" (e.g. to be supported through the undergraduate scholars program) and graduate students (likely to be supported by the NSF AI Institute in Dynamic Systems) --- but is open to others as well. Regular attendance and active participation is expected, to maximize the benefit and learning experience for all.
Spring 2024 Seminars
Beginning January 26, TAILS will take place on Friday afternoons from 4 to 5pm in Wilson Hall 1-119.
Interested?
Email John Smith or Bree Cummins to get added to the email list!
Projects
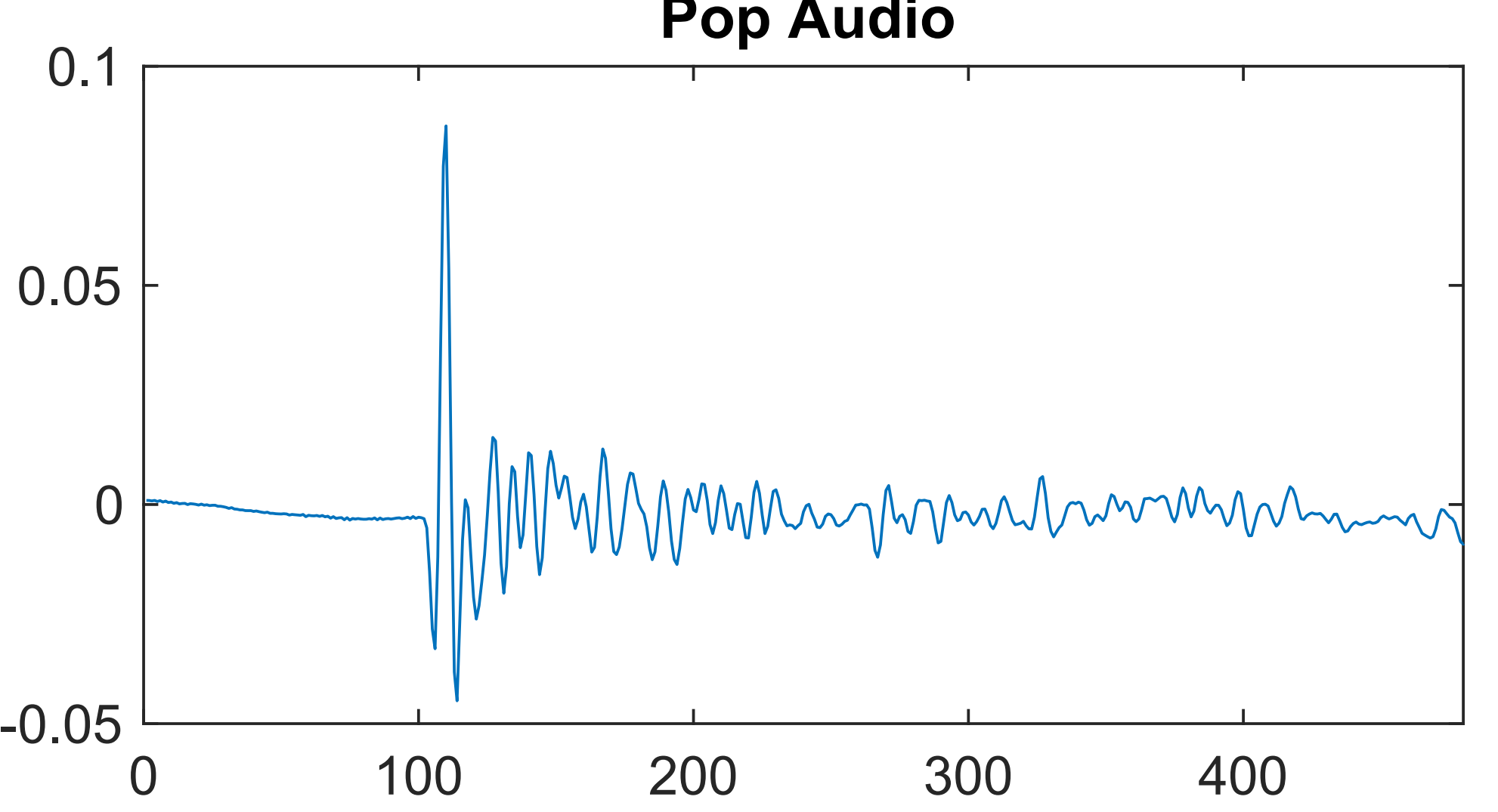
Audial Detection of Leafy Spurge Seed Dispersal
Students : Nat Sweeney, Trevor Vannoy, Brady Hislop
PI's : Dominique Zosso, Brad Whitaker, Katharine Banner
Leafy Spurge (Euphorbia virgata) is an invasive species to Montana. The pop detection project was intended to explore ways to detect the audible "pops" made by its exploding seed pods. Students useda wavelet scattering transform as inputs to an autoencoder. A public github repo for the Leafy Spurge audial detection project is available here.

SET: Computer Vision
Student : Zac Phaneuf
PI : Dominique Zosso
The card game, SET, consists of 81 unique cards that vary in four features across three possibilities for each kind of feature. The computer vision project aims to create a neural net capable of identifying the four characteristc features of a given card from a photo. Future work aims to expand on the identification of cards to the identification of valid sets.
Synthetic Biology Error Detection
Student: Ben Vogel
PI's : Breschine Cummins, Dominique Zosso
Flow cytometry fluorescence data were collected to evaluate the performance of a synthetic biology logic circuit embedded in the yeast genome (Cummins et al., 2023). The analysis is dependent on well-separated normal (Gaussian) distributions of low and high fluorescence, but the data, including the controls, were found to be bimodal. This project aims to improve the separation using a Gaussian mixture model (GMM) to separate the bimodal distributions of the control groups and ultimately classify the experimental conditions. Pipette Image Source
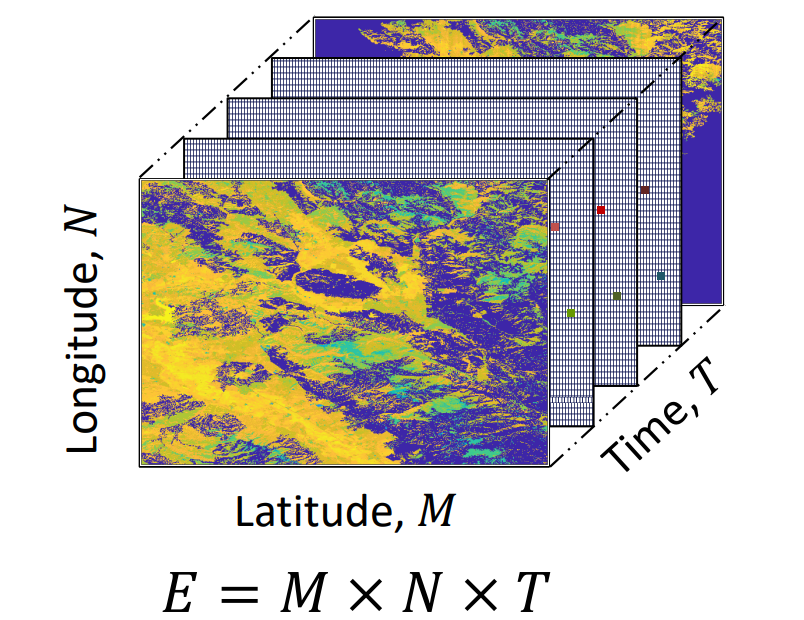
Estimation of Plant Evapotranspiration on a Spatiotemporal Scale Using Low-Rank Tensor Factorization
Students: Farshina Nazrul Shimim, Jacob Munson
PI's: Dominique Zosso, Gaurav Jha, Bradley Whitaker
This project aims to predict high-resolution spatial Evapotranspiration (ET) data within an agricultural field by addressing both temporal and spatial information gaps. Three in-field sensors provide daily ET data at specific locations within the field, while weekly/biweekly aerial missions capture spatial ET data but only at discrete time points. We seek to fill spatio-temporal gaps using statistical and machine learning methodologies.To accomplish this objective, we employ a low-rank tensor approximation framework to model the spatiotemporal ET data. Leveraging the available data, we apply various solvers, including Gradient Descent, BiasMF, FunkSVD, Genetic Algorithm, among others, to predict missing values within the modeled spatiotemporal ET matrix.
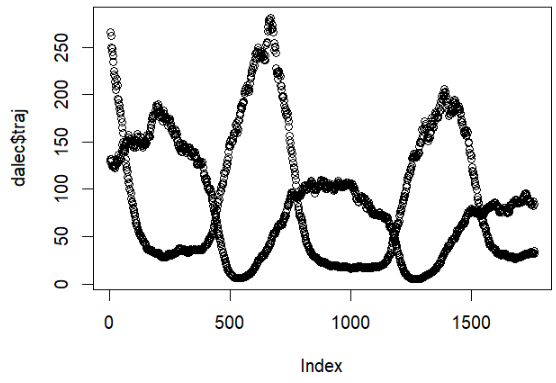
LogLAI : R Package
Student: Turner Haugen
PI: John Smith
LogLAI is an R package that is currently in progress! It contains functions created by John Smith that use Lognormal State Space Functions to project leaf area index and carbon sources in forests. These commands take in synthetic/real data, use them to create particle filters, and these particle filters are then used to create objective functions, which contain marginal loglikelihood and trajectory of the carbon sources we were keeping track of. A large amount of these commands were built in pomp, which lets us interface particle filter implementations in C, making them run faster! The LogLAI package can be accessed here and can be installed in R after installing devtools with the command: install_github("turnerch/loglai")
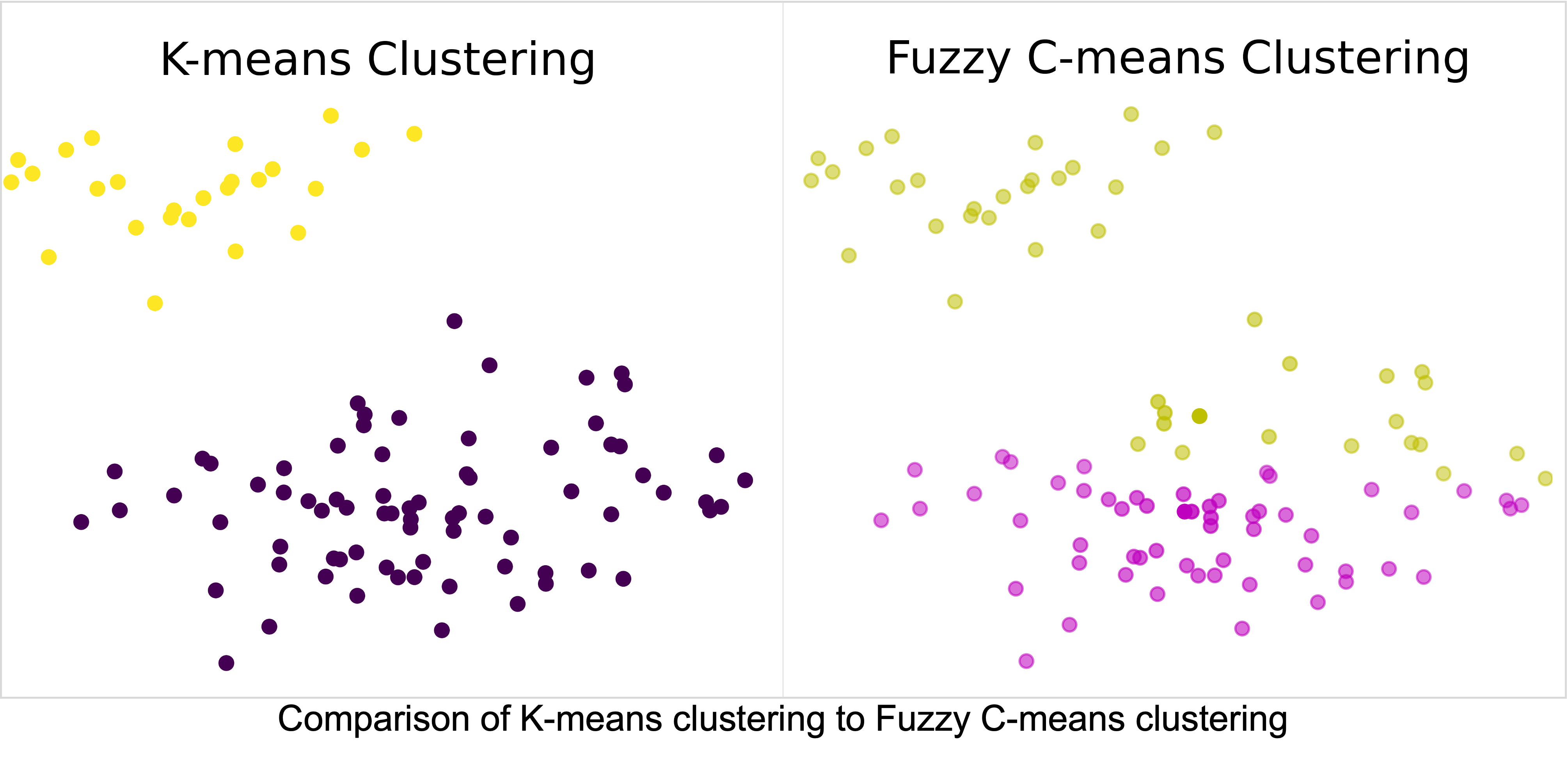
Comparison of Fuzzy C-Means, K-Means and Gaussian Mixture Models
Student: Brady Hislop
Clustering is a common unsupervised machine learning approach that looks to identify groups within unlabeled data. The identification of groups is a critical aspect of biological studies that are used to identify potential biomarkers of disease. These potential biomarkers of disease are often used to drive future studies of a disease, thus, performing valid clustering of the data is critical. In this study, we investigate three clustering algorithms fuzzy c-means, k-means, and gaussian mixture models and the impacts of hyperparameter selection on clustering results. Furthermore, we will demonstrate how the application of each of these algorithms impacts the discovered clusters within biological data, and ultimately impacts future biological directions.
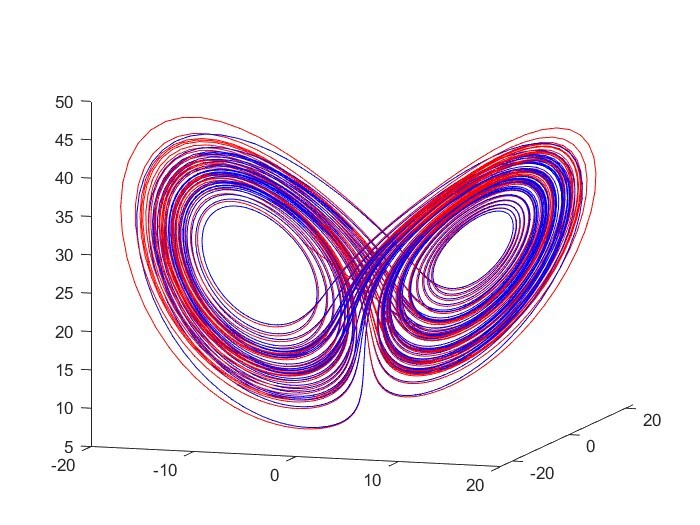
Identifying Physical Laws from Data
Student : Derek Jollie
PI: Scott McCalla
This project aims to use various machine learning algorithms specialized in identifying nonlinear dynamics such as SINDy, sparse model selection via integral terms, and a neural net, to find models from data. One goal is to model the friction forces on projectiles moving through air. A second goal is to build a forced Van der Pol circuit and identify a working model through the algorithms.
